Kinetochore-Biology
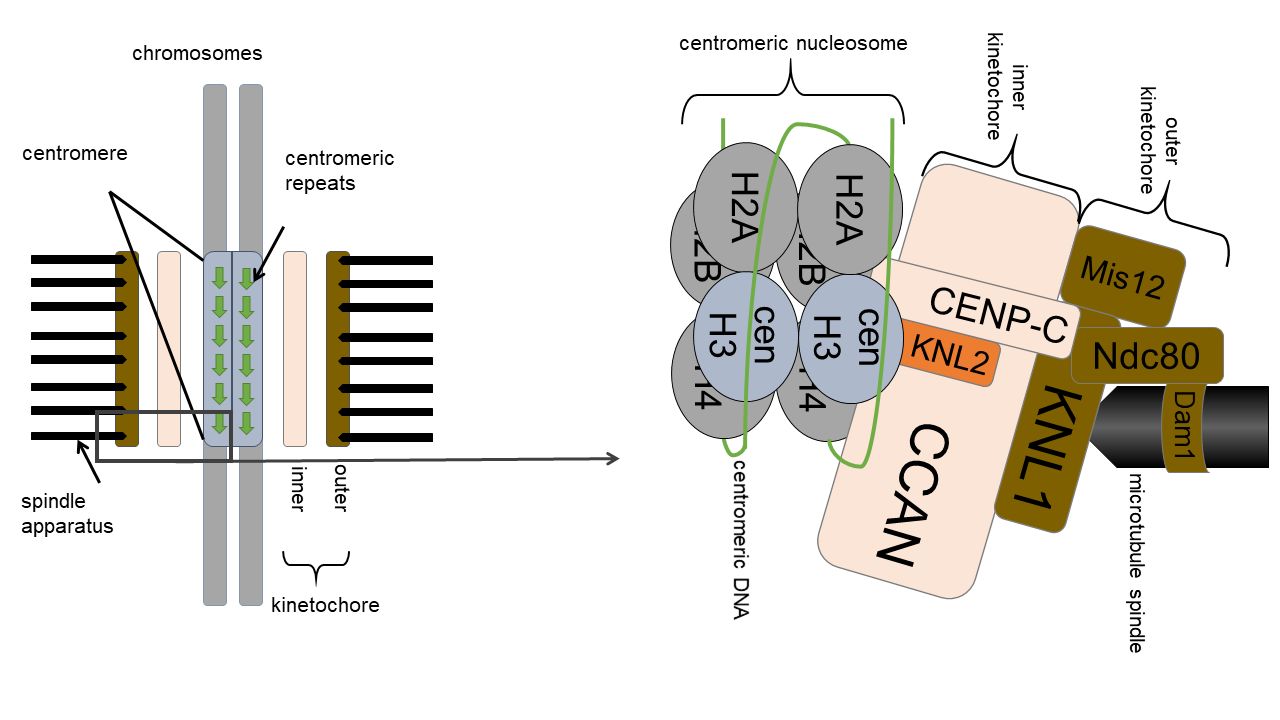
The kinetochore is located on the centromere, where tandemly arranged centromeric repeats form a compact higher-order structure. The kinetochore is a multimeric protein complex with diverse interactions between its compounds, as illustrated by the schematic model.
The research group Kinetochore Biology (KB) is interested in studying the organisation, establishment and maintenance of the kinetochore complex in plants.
The kinetochore is a large protein complex (up to 100 proteins) that assembles at specialised chromosomal domains, the centromeres, during cell divisions. The main function of this complex is to ensure the proper segregation of chromosomes during mitosis and meiosis and, consequently, genome stability in all eukaryotic organisms.
The centromere-specific histone H3-variant CenH3 is essential for kinetochore formation and centromere function. Its deposition at centromeres depends on CenH3 assembly factors, chaperones, transcription of the centromeric repeats and the epigenetic status of centromeric chromatin. Specific manipulation of the CenH3 assembly factor KNL2 yielded double haploids in Arabidopsisthaliana (I. Lermontova, WO2017/067714). The production of double haploids enables a shortcut to achieve genome homozygosity in plant breeding because it allows to produce homogeneous lines in one rather than five or more generations as in conventional breeding.
scroll top
Projects
Our main research goals are:
1. To elucidate the mechanism of kinetochore assembly and function in plants.
We focus on:
- Deciphering the centromere-specific histone H3-variant (cenH3) loading mechanism in plants.
- Identification and functional characterization of novel components of the plant kinetochore complex.
- Functional analysis of protein-protein interaction networks of kinetochore components.
- Elucidation of the mechanism of epigenetic regulation in kinetochore assembly.
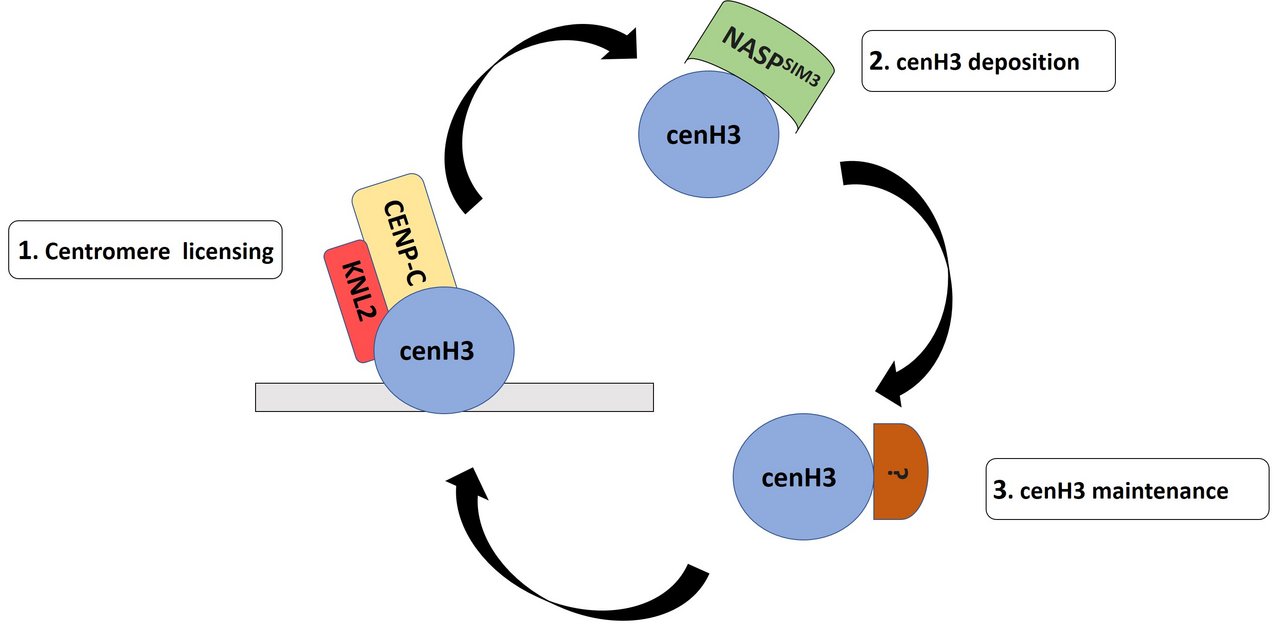
cenH3 loading pathway
In most eukaryotes, normal kinetochore formation depends on the centromeric variant of histone H3 cenH3. The cenH3 deposition pathway in A. thaliana is represented by a three-step process: initiation, deposition and maintenance. (1) Initiation of cenH3 incorporation involves the licensing factor Kinetochore Null 2 (KNL2) and other components (Lermontova et al., 2013), (2) then cenH3 is deposited to centromeres by chaperone proteins such as NASPSIM3 (Le Goff et al., 2020). (3) The specific mechanisms/factors for maintaining newly incorporated cenH3 are not yet known.
2. To establish an efficient haploid induction approach based on the manipulation of kinetochore proteins.
We focus on:
- Optimization of KNL2-based haploid induction approach in Arabidopsis and determination of its mechanism.
- Transferring the KNL2-based haploid induction approach to crop species.
Generation of haploids and double haploids based on KNL2 manipulations
Crossing of A. thaliana mutant for the kinetochore protein KNL2 with wild-type results in the generation of haploids. (Lermontova, I.: Generation of haploid plants based on KNL 2. (IPK-Anmeldung), Veröffentlichung: 27.04.2017, IPK-Nr. 2015/01. WO2017/067714 (2017)).
3. To analyse the effect of environmental factors on the kinetochore assembly.
We focus on:
- Studying the effects of environmental conditions on heterochromatin structure, kinetochore assembly and haploid induction efficiency.
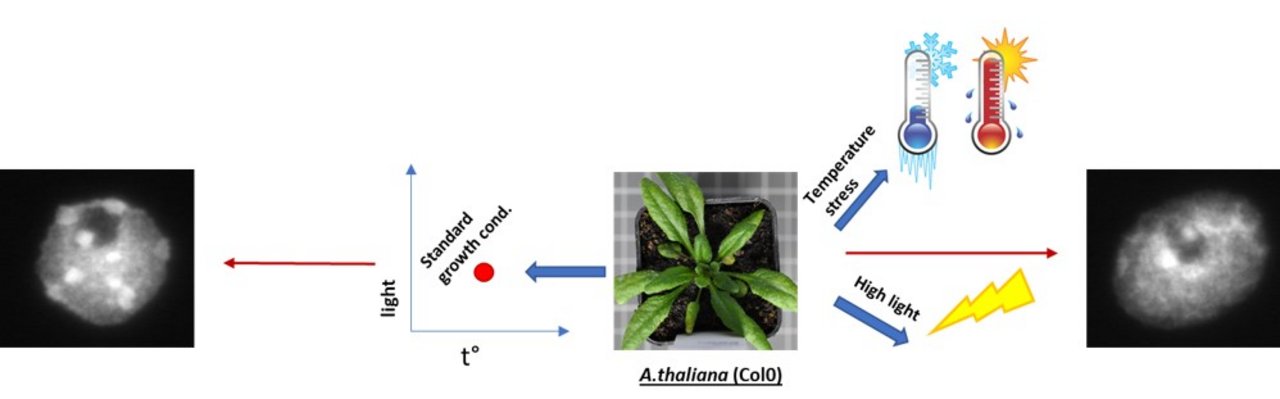
Application of temperature and light stress affect the structure of heterochromatin
Stress conditions such as high temperature and light displayed dispersed heterochromatin structure in A. thaliana.
We apply a combination of molecular biological, biochemical, physiological, cytological, and bioinformatics tools and techniques to achieve our research goals.
scroll top
Staff
scroll top
Publications
Kalidass M, Jarubula V G, Ratnikava M, Chandra J R, Le Goff S, Probst A V, Esposito S, Grasser K D, Bruckmann A, Gagneux J F, Prosée R F, Rutten T, Schubert V, Demidov D, Lechner E, Steiner F A, Genschik P, Lermontova I:
Ubiquitin-dependent proteolysis of KNL2 driven by APC/CCDC20 is critical for centromere integrity and mitotic fidelity. Plant Cell 37 (2025) koaf164. https://dx.doi.org/10.1093/plcell/koaf164
Kalidass M, Vaculíková J, Chandra J R, Králová B, Jarubula V G, Kara Öztürk S D, Demidov D, Schubert V, Potesil D, Palecek J J, Lermontova I:
The C-terminal SUMOylation-dependent regulation of αKNL2 governs its centromere targeting and interaction with CENH3. Plant Commun. (2025) Epub ahead of print. https://dx.doi.org/10.1016/j.xplc.2025.101617
Majolo Willers H:
Functional characterization of RNA polymerases IV and V in the transcriptional regulation of centromeric repeats in Arabidopsis thaliana. (Bachelor Thesis) Berlin, Berliner Hochschule für Technik, Studiengang Biotechnologie (2025) 61 pp.
Yadala R:
Identification and characterization of EMB1674, as a novel KNL2 variant: revealing its centromere targeting mechanism and critical role in plant-specific kinetochore assembly. (PhD Thesis) Halle/S., Martin-Luther-Universität Halle-Wittenberg, Naturwissenschaftliche Fakultät III Agrar- und Ernährungswissenschaften, Geowissenschaften und Informatik (2025) 124 pp.
Yalagapati S P, Ahmadli U, Sinha A, Kalidass M, Dabravolski S, Zuo S, Yadala R, Rutten T, Talbert P, Berr A, Lermontova I:
Centromeric localization of αKNL2 and CENP-C proteins in plants depends on their centromere-targeting domain and DNA-binding regions. Nucleic Acids Res. 53 (2025) gkae1242. https://dx.doi.org/10.1093/nar/gkae1242
Crhak Khaitova L, Mikulkova P, Pecinkova J, Kalidass M, Heckmann S, Lermontova I, Riha K:
Heat stress impairs centromere structure and segregation of meiotic chromosomes in Arabidopsis. eLife 12 (2024) e90253. https://dx.doi.org/10.7554/eLife.90253
Ramakrishnan Chandra J, Kalidass M, Demidov D, Dabravolski S A, Lermontova I:
The role of centromeric repeats and transcripts in kinetochore assembly and function. Plant J. 118 (2024) 982-996. https://dx.doi.org/10.1111/tpj.16445
Yalagapati S:
Centromere targeting mechanism of KNL2 protein variants through analysis of their interaction with centromeric DNA. (Master Thesis) Guntur, India, Biotechnology at Vignans Foundation for Science, Technology & Research (2024) 58 pp.
Ahmadli U, Kalidass M, Crhak Khaitova L, Fuchs J, Cuacos M, Demidov D, Zuo S, Pecinkova J, Mascher M, Ingouff M, Heckmann S, Houben A, Riha K, Lermontova I:
High temperature increases centromere-mediated genome elimination frequency and enhances haploid induction in Arabidopsis. Plant Commun. 4 (2023) 100507. https://dx.doi.org/10.1016/j.xplc.2022.100507
Jarubula V G:
Interaction mapping analysis of E3 ubiquitin ligases with Kinetochore Null2 in Arabidopsis thaliana. (Master Thesis) Köthen, Hochschule Anhalt, Fachbereich Angewandte Biowissenschaften und Prozesstechnik (2023) 44 pp.
Kuo Y-T, Câmara A S, Schubert V, Neumann P, Macas J, Melzer M, Chen J, Fuchs J, Abel S, Klocke E, Huettel B, Himmelbach A, Demidov D, Dunemann F, Mascher M, Ishii T, Marques A, Houben A:
Holocentromeres can consist of merely a few megabase-sized satellite arrays. Nat. Commun. 14 (2023) 3502. https://dx.doi.org/10.1038/s41467-023-38922-7
Schönau L:
Analyse der CENH3 Expression in verschiedenen Sorten von B. napus. (Bachelor Thesis) Jena, Ernst-Abbe-Hochschule Jena, Fachbereich Medizintechnik und Biotechnologie, Studiengang Biotechnologie (2023) 41 pp.
Schubert V, Weißleder A, Lermontova I:
Simultaneous EYFP-CENH3/H2B-DsRed expression is impaired differentially in meristematic and differentiated nuclei of Arabidopsis double transformants. Cytogenet. Genome Res. 163 (2023) 74-80. https://dx.doi.org/10.1159/000533317
Ahmadli U, Sandmann M, Fuchs J, Lermontova I:
Immunolabeling of nuclei/chromosomes in Arabidopsis thaliana. In: Caillaud M-C (Ed.): Plant cell division: methods and protocols, 2nd. ed. (Series: Methods in molecular biology, Vol. 2382) New York [u.a.]: Springer US (2022) ISBN 978-1-0716-1743-4, 19-28. https://dx.doi.org/10.1007/978-1-0716-1744-1_2
Demidov D, Lermontova I, Moebes M, Kochevenko A, Fuchs J, Weiss O, Rutten T, Sorge E, Zuljan E, Giehl R F H, Mascher M, Somasundaram S, Conrad U, Houben A:
Haploid induction by nanobody targeted ubiquitin-proteasome-based degradation of EYFP-tagged CENH3 in Arabidopsis thaliana. J. Exp. Bot. 73 (2022) 7243–7254. https://dx.doi.org/10.1093/jxb/erac359
Möbes M:
Optimierung einer quantitativen Reverse-Transkriptase-PCR Methode zur Analyse niedrig-exprimierter Gene in Nutzpflanzen, basierend auf B. napus. (Bachelor Thesis) Köthen, Hochschule Anhalt, Fachbereich Angewandte Biowissenschaften und Prozesstechnik (2022) 48 pp.
Moebes M, Kuhlmann H, Demidov D, Lermontova I:
Optimization of quantitative reverse transcription PCR method for analysis of weakly expressed genes in crops based on rapeseed. Front. Plant Sci. 13 (2022) 954976. https://dx.doi.org/10.3389/fpls.2022.954976
Sinha A:
The CENPC-k and CENPC motif dependent targeting of proteins to the centromere using EYFP as a marker. (Master Thesis) Kiel, Christian-Albrechts-Universität zu Kiel (2022) 56 pp.
Yadala R, Ratnikava M, Lermontova I:
Bimolecular fluorescence complementation to test for protein-protein interactions and to uncover regulatory mechanisms during gametogenesis. In: Lambing C (Ed.): Plant gametogenesis: methods and protocols, 1st. ed. (Series: Methods in molecular biology, Vol. 2484) New York [u.a.]: Humana Press (2022) ISBN 978-1-0716-2252-0, 107-120. https://dx.doi.org/10.1007/978-1-0716-2253-7_9
Zuo S:
Plastome, repeatome and kinetochore protein evolution in land plants. (PhD Thesis) Brno, Masaryk University (2022) 176 pp.
Zuo S, Yadala R, Yang F, Talbert P, Fuchs J, Schubert V, Ahmadli U, Rutten T, Pecinka A, Lysak M A, Lermontova I:
Recurrent plant-specific duplications of KNL2 and its conserved function as a kinetochore assembly factor. Mol. Biol. Evol. 39 (2022) msac123. https://dx.doi.org/10.1093/molbev/msac123
Capitao C, Tanasa S, Fulnecek J, Raxwal V K, Akimcheva S, Bulankova P, Mikulkova P, Khaitova L C, Kalidass M, Lermontova I, Scheid O M, Riha K:
A CENH3 mutation promotes meiotic exit and restores fertility in SMG7-deficient Arabidopsis. PLoS Genet. 17 (2021) e1009779. https://dx.doi.org/10.1371/journal.pgen.1009779
Houben A, Demidov D, Lermontova I, Fuchs J, Weiss O, Meier K:
Generation of haploids based on mutation of SAD2. (Patent), Veröffentlichung: 24.03.2021, IPK-Nr. 2018/06. EP3794939. (2021).
Municio C, Antosz W, Grasser K, Kornobis E, van Bel M, Eguinoa I, Coppens F, Bräutigam A, Lermontova I, Bruckmann A, Zelkowska K, Houben A, Schubert V:
The Arabidopsis condensin CAP-D subunits arrange interphase chromatin. New Phytol. 230 (2021) 972-987. https://dx.doi.org/10.1111/nph.17221
Sorge E, Demidov D, Lermontova I, Houben A, Conrad U:
Engineered degradation of EYFP-tagged CENH3 via the 26S proteasome pathway in plants. PLoS One 16 (2021) e0247015. https://dx.doi.org/10.1371/journal.pone.0247015
Dvořák Tomaštíková E, Rutten T, Dvořák P, Tugai A, Ptošková K, Petrovská B, van Damme D, Houben A, Doležel J, Demidov D:
Functional divergence of microtubule-associated TPX2 family members in Arabidopsis thaliana. Int. J. Mol. Sci. 21 (2020) 2183. https://dx.doi.org/10.3390/ijms21062183
Ishii T, Juranić M, Maheshwari S, Bustamante F O, Vogt M, Salinas-Gamboa R, Dreissig S, Gursanscky N, How T, Demidov D, Fuchs J, Schubert V, Spriggs A, Vielle-Calzada J P, Comai L, Koltunow A M G, Houben A:
Unequal contribution of two paralogous CENH3 variants in cowpea centromere function. Commun. Biol. 3 (2020) 775. https://dx.doi.org://10.1038/s42003-020-01507-x
Le Goff S, Keçeli B N, Jeřábková H, Heckmann S, Rutten T, Cotterell S, Schubert V, Roitinger E, Mechtler K, Franklin F C H, Tatout C, Houben A, Geelen D, Probst A V, Lermontova I:
The H3 histone chaperone NASPSIM3 escorts CenH3 in Arabidopsis. Plant J. 101 (2020) 71-86. https://dx.doi.org/10.1111/tpj.14518
Boudichevskaia A, Houben A, Fiebig A, Prochazkova K, Pecinka A, Lermontova I:
Depletion of KNL2 results in altered expression of genes involved in regulation of the cell cycle, transcription, and development in Arabidopsis. Int. J. Mol. Sci. 20 (2019) 5726. https://dx.doi.org/10.3390/ijms20225726
Demidov D, Heckmann S, Weiss O, Rutten T, Tomaštíková E D, Kuhlmann M, Scholl P, Municio C M, Lermontova I, Houben A:
Deregulated phosphorylation of CENH3 at Ser65 affects the development of floral meristems in Arabidopsis thaliana. Front. Plant Sci. 10 (2019) 928. https://dx.doi.org/10.3389/Fpls.2019.00928
Kalinowska K, Chamas S, Unkel K, Demidov D, Lermontova I, Dresselhaus T, Kumlehn J, Dunemann F, Houben A:
State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 132 (2019) 593–605. https://dx.doi.org/10.1007/s00122-018-3261-9
Zelkowski M, Zelkowska K, Conrad U, Hesse S, Lermontova I, Marzec M, Meister A, Houben A, Schubert V:
Arabidopsis NSE4 proteins act in somatic nuclei and meiosis to ensure plant viability and fertility. Front. Plant Sci. 10 (2019) 774. https://dx.doi.org/10.3389/fpls.2019.00774
scroll top