Automated Plant Phenotyping
Our central task is the management and coordination of the plant phenotyping infrastructure at IPK, in cooperation with internal and external partners we therefore contribute to the elucidation of plant growth and its plasticity under diverse environmental conditions. We work closely with the Research Group Bioinformatics and Information Technology (BIT) to implement a data management system for standardised documentation, sustainable storage and dissemination of phenomics datasets according to FAIR principles.
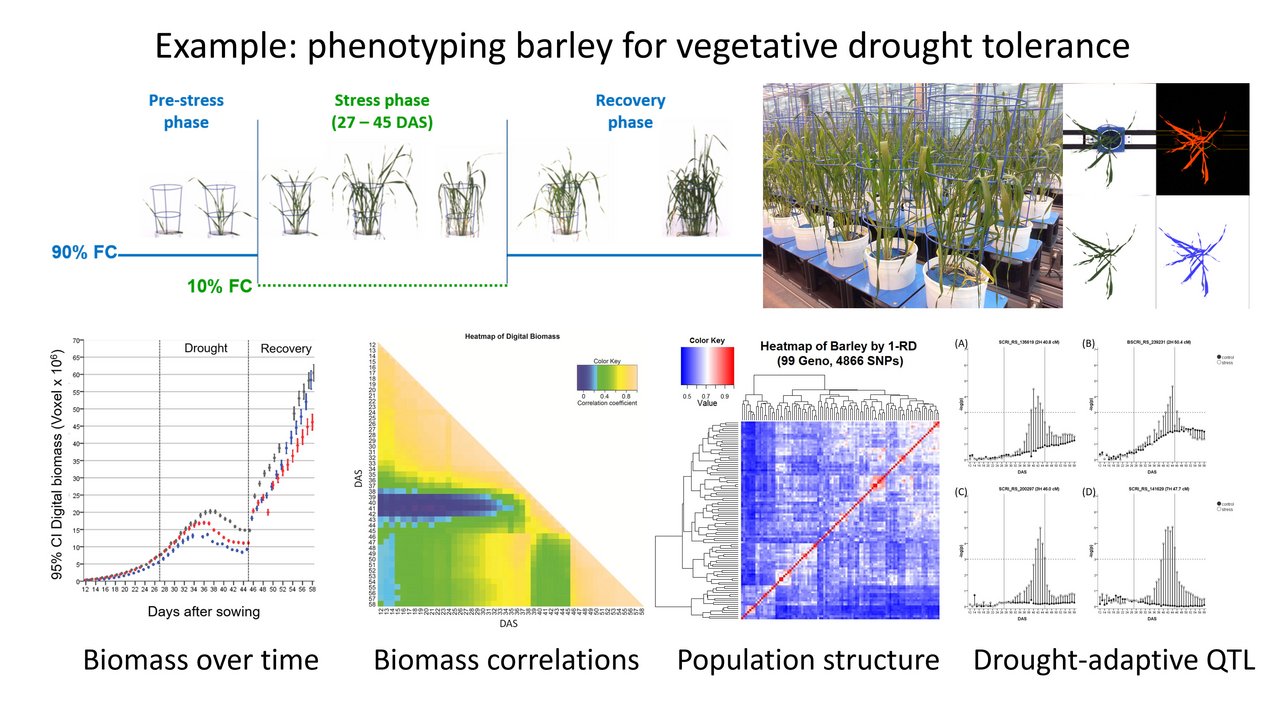
The focus of our own research is predominantly to study the plant response to abiotic stress, especially drought and heat. As this response is very environment-dependent dynamic, we use precision phenotyping systems that allow non-invasive observation under controlled environmental conditions. Our focus is on genetics and physiology and also on modelling plant growth and water use (Fig. 1). Plant materials are mainly plant genetic resources of cereals and legumes, which represent a valuable source for improving the stress tolerance of modern varieties via breeding approaches for the necessary adaptation to the ongoing climate change.
scroll top
Projects
Within the German Plant Phenotyping Network (DPPN), we are expanding the existing infrastructure for plant phenotyping and making it available to users from research and industry within the framework of collaborative projects and transnational access projects (EPPN2020).
STARGATE
Within the scope of STARGATE we will support our partner in Portugal (UCP) to become an established centre of excellence on the use of sensors, multi-omics and plant phenotyping technologies.
BRACE
In BRACE we are studying mechanisms of sustainability under abiotic stresses in the first wild barley multi-parental nested association mapping (NAM) population (HEB-25) using modern automated phenotyping approaches.
BOLERO
Root development of coffee and cocoa plants is the focus of BOLERO, where we use automatic phenotyping to investigate the dynamics of root development and the response to different nutrient supply.
INCREASE
In the frame of INCREASE we are contributing to foster agricultural biodiversity in Europe by evaluating growth and drought tolerance of 200 genetic resources of chickpea via precision phenotyping and by conducting a citizen science experiment across Europe with 1,000 genetic resources of common bean. For three years, citizens throughout Europe have been called upon to participate in the bean diversity experiment. The IPK invited 100 German participants to a Citizens' Day at the institute on July 21, 2023.
Citizen Science Day 2023
scroll top
Staff
scroll top
Publications
Bienert M D, Junker A, Melzer M, Altmann T, von Wirén N, Bienert G P:
Boron deficiency responses in maize (Zea mays L.) roots. J. Plant Nutr. Soil Sci. (2026) Epub ahead of print. https://doi.org/10.1002/jpln.202300173
Laura T, Rosaria T M, Silvia R, Tea S, Teresa A M, Lorenzo B, Luciana G, Salar S, Laura R, Leonardo R G:
Construction of a living library of Solanum tomentosum introgression lines into the S. melongena genome: a tool to exploit novel genetic diversity for eggplant breeding. Hortic. Plant J. (2026) Epub ahead of print. https://doi.org/10.1016/j.hpj.2025.04.019
Narisetti N, Neumann K, Altmann T, Stolzenburg F, von Wirén N, Giehl R F, Gladilin E:
A multi-class deep learning segmentation approach for automated analysis of axial and lateral roots in barley plants. Comput. Electron. Agric. 240 (2026) 111157. https://dx.doi.org/10.1016/j.compag.2025.111157
Alcock T D, Bienert M D, Junker A, Meyer R C, Tschiersch H, Kudamala S, von Wirén N, Altmann T, Bienert G P:
Arabidopsis thaliana exhibits wide within-species variation in tolerance to boron limitation and root and shoot trait resilience associate with a pleiotropic locus. New Phytol. 248 (2025) 2444-2465. https://doi.org/10.1111/nph.70570
Fernández-Alonso M J, de Celis M, Belda I, Palomino J, García C, Gaitán J, Wang J-T, Abdala-Roberts L, Alfaro F D, Angulo-Pérez D F, Arthikala M-K, Chalasani D, Corwin J, Gui-Lan D, Hernandez-Lopez A, Nanjareddy K, Nayaka S C, Pasari B, Patro T S S K, Podile A R, Quijano-Medina T, Rivera D S, Sarma P V S R N, Shaaf S, Trivedi P, Yang Q, Yin Y, Zaady E, Zhu Y-G, Singh B K, Delgado-Baquerizo M, García-Palacios P, Milla R:
Native edaphoclimatic regions shape soil communities of crop wild progenitors. ISME Commun. 5 (2025) ycaf143. https://dx.doi.org/10.1093/ismeco/ycaf143
Gladilin E, Narisetti N, Neumann K, Altmann T:
End-to-end deep learning approach to automated phenotyping of greenhouse-grown plant shoots. Agronomy 15 (2025) 1117. https://dx.doi.org/10.3390/agronomy15051117
Inoue M, Situmorang A, Kelly J H, Chen W, Zhou H, Ferrario C C, Gregis V, Vajani A, Shaaf S, Biswas A, Alqusumi R, Waters M T, Tucker M R, Zhang D, Watts-Williams S J, Janiak A, Marzec M, Chmielewska B, Rossini L, Yoneyama K, Brewer P B:
LATERAL BRANCHING OXIDOREDUCTASE specificity for strigolactone branching inhibition in barley. J. Exp. Bot. 76 (2025) 5367-5381. https://dx.doi.org/10.1093/jxb/eraf285
Lauterberg M M:
Utilizing high-throughput phenotyping to dissect plant genetic resources of wheat and chickpea under drought stress. (PhD Thesis) Halle/S., Martin-Luther-Universität Halle-Wittenberg, Naturwissenschaftliche Fakultät III Agrar- und Ernährungswissenschaften, Geowissenschaften und Informatik (2025) 101 pp.
Montardit-Tarda F, Casas A M, Thomas W T B, Schnaithmann F, Sharma R, Shaaf S, Campoli C, Russell J, Ramsay L, Bayer M M, Delbono S, Jääskeläinen M, Paul M, Stoddard F L, Visioni A, Flavell A J, Pillen K, Kilian B, Graner A, Rossini L, Waugh R, Cattivelli L, Schulman A H, Tondelli A, Igartua E:
New loci and candidate genes in spring two-rowed barley detected through meta-analysis of a field trial European network. Theor. Appl. Genet. 138 (2025) 158. https://dx.doi.org/10.1007/s00122-025-04934-8
Sahu A, Psaroudakis D, Rolletschek H, Neumann K, Borisjuk L, Himmelbach A, Pinninti K, Knoch D, Töpfer N, Szymanski J:
panomiX: Investigating mechanisms of trait emergence through multi-omics data integration. Plant Phenomics 7 (2025) 100131. https://doi.org/10.1016/j.plaphe.2025.100131
Schüler D, Lange M, Altmann T, Cuacos M, Arend D, D’Auria J C, Fiebig A, Kumlehn J, Neumann K, Melzer M, Rey-Mazón E, Rolletschek H, Scholz U, Willner E, Reif J C:
Data management in balance – a decade of balancing pragmatism, sustainability and innovation at plant research center IPK Gatersleben. J. Integr. Bioinform. 22 (2025) 20250012. https://dx.doi.org/10.1515/jib-2025-0012
Sharma R, Shaaf S, Neumann K, Civan P, Guo Y, Mascher M, David M, Al-Yassin A, Özkan H, Blake T, Hübner S, Castañeda-Álvarez N P, Grando S, Ceccarelli S, Baum M, Graner A, Coupland G, Pillen K, Weiss E, Mackay I J, Powell W, Kilian B:
On the origin of the late-flowering ppd-H1 allele in barley. Theor. Appl. Genet. 138 (2025) 246. https://dx.doi.org/10.1007/s00122-025-04981-1
Shi R, López-Malvar A, Knoch D, Tschiersch H, Heuermann M C, Shaaf S, Madur D, Santiago R, Balconi C, Frascaroli E, Erdal S, Palaffre C, Charcosset A, Revilla P, Altmann T:
Integrating high-throughput phenotyping and genome-wide association analyses to unravel Mediterranean maize resilience to combined drought and high temperatures. Plant Stress 17 (2025) 100954. https://doi.org/10.1016/j.stress.2025.100954
Shi R, López-Malvar A, Knoch D, Tschiersch H, Heuermann M C, Shaaf S, Madur D, Santiago R, Balconi C, Frascaroli E, Erdal S, Palaffre C, Charcosset A, Revilla P, Altmann T:
A high-throughput phenotyping dataset for GWAS analysis of maize under combined drought and heat stress. Data Brief 62 (2025) 111947. https://dx.doi.org/10.1016/j.dib.2025.111947
Soleimani S, Wichmann P, Pillen K, Neumann K, Maurer A:
Tillering plasticity of drought-stressed barley genotypes under different re-watering regimes. BMC Plant Biol. 25 (2025) 1368. https://dx.doi.org//10.1186/s12870-025-07504-8
Turek S, Skarzyńska-Łyżwa A, Aparna A, Pląder W, Riewe D, Junker A, Altmann T, Pawełkowicz M:
Multi-omics integration of transcriptome, miRNA, and metabolome uncovers molecular mechanisms of male flower development in cucumber line B10 (Cucumis sativus L.). Sci. Rep. 15 (2025) 45734. https://dx.doi.org/10.1038/s41598-025-28485-6
Ullah S, Narisetti N, Neumann K, Altmann T, Hejatko J, Gladilin E:
Automated generation of ground truth images of greenhouse-grown plant shoots using a GAN approach. Plant Methods 21 (2025) 126. https://dx.doi.org/10.1186/s13007-025-01441-1
Wonneberger R, DAuria J C, Neumann K, Hansen P B, Dieseth J A, Nielsen L K, Niemelä T, Odilbekov F, Novakazi F, Bengtsson T, CResWheat Consortium:
Integrating metabolomics and high-throughput phenotyping to elucidate metabolic and phenotypic responses to early-season drought stress in Nordic spring wheat. BMC Plant Biol. 25 (2025) 987. https://dx.doi.org/10.1186/s12870-025-06914-y
Cortinovis G, Vincenzi L, Anderson R, Marturano G, Marsh J I, Bayer P E, Rocchetti L, Frascarelli G, Lanzavecchia G, Pieri A, Benazzo A, Bellucci E, Di Vittori V, Nanni L, Ferreira Fernández J J, Rossato M, Aguilar O M, Morrell P L, Rodriguez M, Gioia T, Neumann K, Alvarez Diaz J C, Gratias A, Klopp C, Bitocchi E, Geffroy V, Delledonne M, Edwards D, Papa R:
Adaptive gene loss in the common bean pan-genome during range expansion and domestication. Nat. Commun. 15 (2024) 6698. https://dx.doi.org/10.1038/s41467-024-51032-2
de Celis M, Fernandéz-Alonso M J, Belda I, García C, Ochoa-Hueso R, Palomino J, Singh B K, Yin Y, Wang J-T, Abdala-Roberts L, Alfaro F D, Angulo-Pérez D, Arthikala M-K, Corwin J, Gui-Lan D, Hernandez-Lopez A, Nanjareddy K, Pasari B, Quijano-Medina T, Rivera D S, Shaaf S, Trivedi P, Yang Q, Zaady E, Zhu Y-G, Delgado-Baquerizo M, Milla R, García-Palacios P:
The abundant fraction of soil microbiomes regulates the rhizosphere function in crop wild progenitors. Ecol. Lett. 27 (2024) e14462. https://dx.doi.org/10.1111/ele.14462
Heuermann M C, Meyer R C, Knoch D, Tschiersch H, Altmann T:
Strong prevalence of light regime-specific QTL in Arabidopsis detected using automated high-throughput phenotyping in fluctuating or constant light. Physiol. Plant. 176 (2024) e14255. https://dx.doi.org/10.1111/ppl.14255
Lauterberg M, Tschiersch H, Zhao Y, Kuhlmann M, Mücke I, Papa R, Bitocchi E, Neumann K:
Implementation of theoretical non-photochemical quenching (NPQ((T))) to investigate NPQ of chickpea under drought stress with High-throughput Phenotyping. Sci. Rep. 14 (2024) 13970. https://dx.doi.org/10.1038/s41598-024-63372-6
Pinninti K:
Transcriptomic analysis of thermomorphogenesis in tomato seedlings. (Master Thesis) Kiel, Christian-Albrechts-Universität zu Kiel (2024)
Ullah S, Panzarová K, Trtílek M, Lexa M, Máčala V, Neumann K, Altmann T, Hejátko J, Pernisová M, Gladilin E:
High-throughput spike detection in greenhouse cultivated grain crops with attention mechanisms-based deep learning models. Plant Phenomics 6 (2024) 0155. https://dx.doi.org/10.34133/plantphenomics.0155
Bellucci E, Benazzo A, Xu C, Bitocchi E, Rodriguez M, Alseekh S, Di Vittori V, Gioia T, Neumann K, Cortinovis G, Frascarelli G, Murube E, Trucchi E, Nanni L, Ariani A, Logozzo G, Shin J H, Liu C, Jiang L, Ferreira J J, Campa A, Attene G, Morrell P L, Bertorelle G, Graner A, Gepts P, Fernie A R, Jackson S A, Papa R:
Selection and adaptive introgression guided the complex evolutionary history of the European common bean. Nat. Commun. 14 (2023) 1908. https://dx.doi.org/10.1038/s41467-023-37332-z
Heuermann M C, Knoch D, Junker A, Altmann T:
Natural plant growth and development achieved in the IPK PhenoSphere by dynamic environment simulation. Nat. Commun. 14 (2023) 5783. https://dx.doi.org/10.1038/s41467-023-41332-4
Khodaeiaminjan M, Knoch D, Ndella Thiaw M R, Marchetti C F, Kořínková N, Techer A, Nguyen T D, Chu J, Bertholomey V, Doridant I, Gantet P, Graner A, Neumann K, Bergougnoux V:
Genome-wide association study in two-row spring barley landraces identifies QTL associated with plantlets root system architecture traits in well-watered and osmotic stress conditions. Front. Plant Sci. 14 (2023) 1125672. https://dx.doi.org/10.3389/fpls.2023.1125672
Lauterberg M, Tschiersch H, Papa R, Bitocchi E, Neumann K:
Engaging precision phenotyping to scrutinize vegetative drought tolerance and recovery in chickpea plant genetic resources. Plants 12 (2023) 2866. https://dx.doi.org/10.3390/plants12152866
Neumann K, Schulthess A W, Bassi F M, Dhanagond S, Khlestkina E, Börner A, Graner A, Kilian B:
Genomic approaches to using diversity for the adaptation of modern varieties of wheat and barley to climate change. In: Ghamkhar K, Williams W, Brown A H D (Eds.): Plant Genetic Resources for the 21st Century. The OMICS Era. New York: Apple Academic Press (2023) ISBN 9781774910825, 47-78. https://dx.doi.org/10.1201/9781003302957
Papalini S, Di Vittori V, Pieri A, Allegrezza M, Frascarelli G, Nanni L, Bitocchi E, Bellucci E, Gioia T, Pereira L G, Susek K, Tenaillon M, Neumann K, Papa R:
Challenges and opportunities behind the use of Herbaria in paleogenomics studies. Plants 12 (2023) 3452. https://dx.doi.org/10.3390/plants12193452
Peterson A, Kishchenko O, Kuhlmann M, Tschiersch H, Fuchs J, Tikhenko N, Schubert I, Nagel M:
Cryopreservation of duckweed genetic diversity as model for long-term preservation of aquatic flowering plants. Plants 12 (2023) 3302. https://dx.doi.org/10.3390/plants12183302
Radchuk V, Belew Z M, Gündel A, Mayer S, Hilo A, Hensel G, Sharma R, Neumann K, Ortleb S, Wagner S, Muszynska A, Crocoll C, Xu D, Hoffie I, Kumlehn J, Fuchs J, Peleke F F, Szymanski J J, Rolletschek H, Nour-Eldin H H, Borisjuk L:
SWEET11b transports both sugar and cytokinin in developing barley grains. Plant Cell 35 (2023) 2186-2207. https://dx.doi.org/10.1093/plcell/koad055
Shi R, Seiler C, Knoch D, Junker A, Altmann T:
Integrated phenotyping of root and shoot growth dynamics in maize reveals specific interaction patterns in inbreds and hybrids and in response to drought. Front. Plant Sci. 14 (2023) 1233553. https://dx.doi.org/10.3389/fpls.2023.1233553
Zahn T, Zhu Z, Ritoff N, Krapf J, Junker A, Altmann T, Schmutzer T, Tüting C, Kastritis P L, Babben S, Quint M, Pillen K, Maurer A:
Novel exotic alleles of EARLY FLOWERING 3 determine plant development in barley. J. Exp. Bot. 74 (2023) 3630-3650. https://dx.doi.org/10.1093/jxb/erad127
Amitrano C, Junker A, DAgostino N, De Pascale S, De Micco V:
Integration of high-throughput phenotyping with anatomical traits of leaves to help understanding lettuce acclimation to a changing environment. Planta 256 (2022) 68. https://dx.doi.org/10.1007/s00425-022-03984-2
Arend D, Psaroudakis D, Memon J A, Rey-Mazón E, Schüler D, Szymanski J J, Scholz U, Junker A, Lange M:
From data to knowledge - big data needs stewardship, a plant phenomics perspective. Plant J. 111 (2022) 335-347. https://dx.doi.org/10.1111/tpj.15804
Badaeva E D, Konovalov F A, Knüpffer H, Fricano A, Ruban A S, Kehel Z, Zoshchuk S A, Surzhikov S A, Neumann K, Graner A, Hammer K, Filatenko A, Bogaard A, Jones G, Özkan H, Kilian B:
Genetic diversity, distribution and domestication history of the neglected GGAtAt genepool of wheat. Theor. Appl. Genet. 135 (2022) 755–776. https://dx.doi.org/10.1007/s00122-021-03912-0
Deblieck M, Szilagyi G, Andrii F, Saranga Y, Lauterberg M, Neumann K, Krugman T, Perovic D, Pillen K, Ordon F:
Dissection of a grain yield QTL from wild emmer wheat reveals sub-intervals associated with culm length and kernel number. Front. Genet. 13 (2022) 955295. https://dx.doi.org/10.3389/fgene.2022.955295
Langstroff A, Heuermann M C, Stahl A, Junker A:
Opportunities and limits of controlled-environment plant phenotyping for climate response traits. Theor. Appl. Genet. 135 (2022) 1–16. https://dx.doi.org/10.1007/s00122-021-03892-1
Lauterberg M, Saranga Y, Deblieck M, Klukas C, Krugman T, Perovic D, Ordon F, Graner A, Neumann K:
Precision phenotyping across the life cycle to validate and decipher drought-adaptive QTLs of wild emmer wheat (Triticum turgidum ssp. dicoccoides) introduced into elite wheat varieties. Front. Plant Sci. 13 (2022) 965287. https://dx.doi.org/10.3389/fpls.2022.965287
Narisetti N, Henke M, Neumann K, Stolzenburg F, Altmann T, Gladilin E:
Deep learning based greenhouse image segmentation and shoot phenotyping (DeepShoot). Front. Plant Sci. 13 (2022) 906410. https://dx.doi.org/10.3389/fpls.2022.906410
Bellucci E, Aguilar O M, Alseekh S, Bett K, Brezeanu C, Cook D, De la Rosa L, Delledonne M, Dostatny D F, Ferreira J J, Geffroy V, Ghitarrini S, Kroc M, Kumar Agrawal S, Logozzo G, Marino M, Mary-Huard T, McClean P, Meglič V, Messer T, Muel F, Nanni L, Neumann K, Servalli F, Străjeru S, Varshney R K, Vasconcelos M W, Zaccardelli M, Zavarzin A, Bitocchi E, Frontoni E, Fernie A R, Gioia T, Graner A, Guasch L, Prochnow L, Opperman M, Susek K, Tenaillon M, Papa R:
The INCREASE project: Intelligent collections of food-legume genetic resources for European agrofood systems. Plant J. 108 (2021) 646-660. https://dx.doi.org/10.1111/tpj.15472
Cortinovis G, Oppermann M, Neumann K, Graner A, Gioia T, Marsella M, Alseekh S, Fernie A R, Papa R, Bellucci E, Bitocchi E:
Towards the development, maintenance, and standardized phenotypic characterization of single-seed-descent genetic resources for common bean. Curr. Protoc. 1 (2021) e133. https://dx.doi.org/10.1002/cpz1.133
Dodig D, Božinović S, Nikolić A, Zorić M, Vančetović J, Ignjatović-Micić D, Delić N, Weigelt-Fischer K, Altmann T, Junker A:
Dynamics of maize vegetative growth and drought adaptability using image-based phenotyping under controlled conditions. Front. Plant Sci. 12 (2021) 652116. https://dx.doi.org/10.3389/fpls.2021.652116
Fadoul H E, Martínez Rivas F J, Neumann K, Balazadeh S, Fernie A R, Alseekh S:
Comparative molecular and metabolic profiling of two contrasting wheat cultivars under drought stress. Int. J. Mol. Sci. 22 (2021) 13287. https://dx.doi.org/10.3390/ijms222413287
Henke M, Neumann K, Altmann T, Gladilin E:
Semi-automated ground truth segmentation and phenotyping of plant structures using k-means clustering of eigen-colors (kmSeg). Agriculture 11 (2021) 1098. https://dx.doi.org/10.3390/agriculture11111098
Kroc M, Tomaszewska M, Czepiel K, Bitocchi E, Oppermann M, Neumann K, Guasch L, Bellucci E, Alseekh S, Graner A, Fernie A R, Papa R, Susek K:
Towards development, maintenance, and standardized phenotypic characterization of single-seed-descent genetic resources for lupins. Curr. Protoc. 1 (2021) e191. https://dx.doi.org/10.1002/cpz1.191
Li M, Hensel G, Melzer M, Junker A, Tschiersch H, Ruwe H, Arend D, Kumlehn J, Börner T, Stein N:
Mutation of the ALBOSTRIANS ohnologous gene HvCMF3 impairs chloroplast development and thylakoid architecture in barley. Front. Plant Sci. 12 (2021) 732608. https://dx.doi.org/10.3389/fpls.2021.732608
Li M, Ruwe H, Melzer M, Junker A, Hensel G, Tschiersch H, Schwenkert S, Chamas S, Schmitz-Linneweber C, Börner T, Stein N:
The Arabidopsis AAC proteins CIL and CIA2 are sub-functionalized paralogs involved in chloroplast development. Front. Plant Sci. 12 (2021) 681375. https://dx.doi.org/10.3389/fpls.2021.681375
Mayer G, Müller W, Schork K, Uszkoreit J, Weidemann A, Wittig U, Rey M, Quast C, Felden J, Glöckner F O, Lange M, Arend D, Beier S, Junker A, Scholz U, Schüler D, Kestler H A, Wibberg D, Pühler A, Twardziok S, Eils J, Eils R, Hoffmann S, Eisenacher M, Turewicz M:
Implementing FAIR data management within the German Network for Bioinformatics Infrastructure (de.NBI) exemplified by selected use cases. Brief. Bioinform. 22 (2021) bbab010. https://dx.doi.org/10.1093/bib/bbab010
Narisetti N, Henke M, Seiler C, Junker A, Ostermann J, Altmann T, Gladilin E:
Fully-automated root image analysis (faRIA). Sci. Rep. 11 (2021) 16047. https://dx.doi.org/10.1038/s41598-021-95480-y
Pommier C, Gruden K, Junker A, Coppens F, Finkers R, Hassani-Pak K, Faria D, Hancock J M, Beier S, Costa B, Miguel C, Chaves I, Davey R, Contreras-Moreira B:
ELIXIR Plant sciences 2020-2023 Roadmap [version 1; not peer reviewed]. F1000Research 10 (2021) 145. https://doi.org/10.7490/f1000research.1118482.1
Rebola-Lichtenberg J, Streit J, Schall P, Ammer C, Seidel D:
From facilitation to competition: the effect of black locust (Robinia pseudoacacia L.) on the growth performance of four poplar-hybrids (Populus spp.) in mixed short rotation coppice. New Forest. 52 (2021) 639-656. https://dx.doi.org/10.1007/s11056-020-09813-2
Sharma S, Schulthess A W, Bassi F M, Badaeva E D, Neumann K, Graner A, Özkan H, Werner P, Knüpffer H, Kilian B:
Introducing beneficial alleles from plant genetic resources into the wheat germplasm. Biology 10 (2021) 982. https://dx.doi.org/10.3390/biology10100982
Arend D, König P, Junker A, Scholz U, Lange M:
The on-premise data sharing infrastructure e!DAL: Foster FAIR data for faster data acquisition. GigaScience 9 (2020) giaa107. https://doi.org/10.1093/gigascience/giaa107
Henke M, Junker A, Neumann K, Altmann T, Gladilin E:
A two-step registration-classification approach to automated segmentation of multimodal images for high-throughput greenhouse plant phenotyping. Plant Methods 16 (2020) 95. https://dx.doi.org/10.1186/s13007-020-00637-x
Mang C:
Characterization of high light acclimation capacity in Arabidopsis mutant candidates. (Master Thesis) Mittweida, Hochschule Mittweida, Fakultät Angewandte Computer- und Biowissenschaften (2020) 81 pp.
Mikołajczak K, Ogrodowicz P, Ćwiek-Kupczyńska H, Weigelt-Fischer K, Mothukuri S R, Junker A, Altmann T, Krystkowiak K, Adamski T, Surma M, Kuczyńska A, Krajewski P:
Image phenotyping of spring barley (Hordeum vulgare L.) RIL population under drought: selection of traits and biological interpretation. Front. Plant Sci. 11 (2020) 743. https://dx.doi.org/10.3389/fpls.2020.00743
Papoutsoglou E A, Faria D, Arend D, Arnaud E, Athanasiadis I N, Chaves I, Coppens F, Cornut G, Costa B V, Ćwiek-Kupczyńska H, Droesbeke B, Finkers R, Gruden K, Junker A, King G J, Krajewski P, Lange M, Laporte M-A, Michotey C, Oppermann M, Ostler R, Poorter H, Ramı́rez-Gonzalez R, Ramšak Ž, Reif J C, Rocca-Serra P, Sansone S-A, Scholz U, Tardieu F, Uauy C, Usadel B, Visser R G F, Weise S, Kersey P J, Miguel C M, Adam-Blondon A-F, Pommier C:
Enabling reusability of plant phenomic datasets with MIAPPE 1.1. New Phytol. 227 (2020) 260-273. https://dx.doi.org/10.1111/nph.16544
Psaroudakis D:
A multi-omics perspective on Arabidopsis drought response: discovering novel regulators with a machine learning based gene-to-phene approach. (Master Thesis) Mittweida, Hochschule Mittweida, Fakultät Angewandte Computer- und Biowissenschaften (2020) 57 pp.
Psaroudakis D, Liu F, König P, Scholz U, Junker A, Lange M, Arend D:
isa4j: a scalable Java library for creating ISA-Tab metadata [version 1; peer review: 2 approved]. F1000Research 9(ELIXIR) (2020) 1388. https://doi.org/10.12688/f1000research.27188.1
Dodig D, Bozinovic S, Nikolic A, Zoric M, Vancetovic J, Ignjatovic-Micic D, Delic N, Weigelt-Fischer K, Junker A, Altmann T:
Image-derived traits related to mid-season growth performance of maize under nitrogen and water stress. Front. Plant Sci. 10 (2019) 814. https://dx.doi.org/10.3389/Fpls.2019.00814
Ghaffar M, Schüler D, König P, Arend D, Junker A, Scholz U, Lange M:
Programmatic access to FAIRified digital plant genetic resources. J. Integr. Bioinform. 16 (2019) 20190060. https://dx.doi.org/10.1515/jib-2019-0060
Henke M, Junker A, Neumann K, Altmann T, Gladilin E:
Comparison of feature point detectors for multimodal image registration in plant phenotyping. PLoS One 14 (2019) e0221203. https://dx.doi.org/10.1371/journal.pone.0221203
Henke M, Junker A, Neumann K, Altmann T, Gladilin E:
Comparison and extension of three methods for automated registration of multimodal plant images. Plant Methods 15 (2019) 44. https://dx.doi.org/10.1186/s13007-019-0426-8
Lobet G, Paez-Garcia A, Schneider H, Junker A, Atkinson J A, Tracy S:
Demystifying roots: A need for clarification and extended concepts in root phenotyping. Plant Sci. 282 (2019) 11-13. https://doi.org/10.1016/j.plantsci.2018.09.015
Narisetti N, Henke M, Seiler C, Shi R, Junker A, Altmann T, Gladilin E:
Semi-automated Root Image Analysis (saRIA). Sci. Rep. 9 (2019) 19674. https://dx.doi.org/10.1038/s41598-019-55876-3
Henke M, Junker A, Neumann K, Altmann T, Gladilin E:
Automated alignment of multi-modal plant images using integrative phase correlation approach. Front. Plant Sci. 9 (2018) 1519. https://dx.doi.org/10.3389/fpls.2018.01519
Pommerrenig B, Junker A, Abreu I, Bieber A, Fuge J, Willner E, Bienert M D, Altmann T, Bienert G P:
Identification of rapeseed (Brassica napus) cultivars with a high tolerance to boron-deficient conditions. Front. Plant Sci. 9 (2018) 1142. https://dx.doi.org/10.3389/fpls.2018.01142
Shi R, Junker A, Seiler C, Altmann T:
Phenotyping roots in darkness: disturbance-free root imaging with near infrared illumination. Funct. Plant Biol. 45 (2018) 400-411. https://doi.org/10.1071/FP17262
scroll top