Research Interest
Head: Prof. Andreas Graner
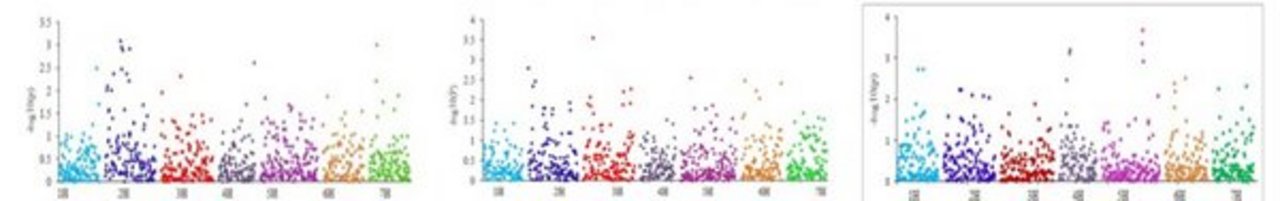
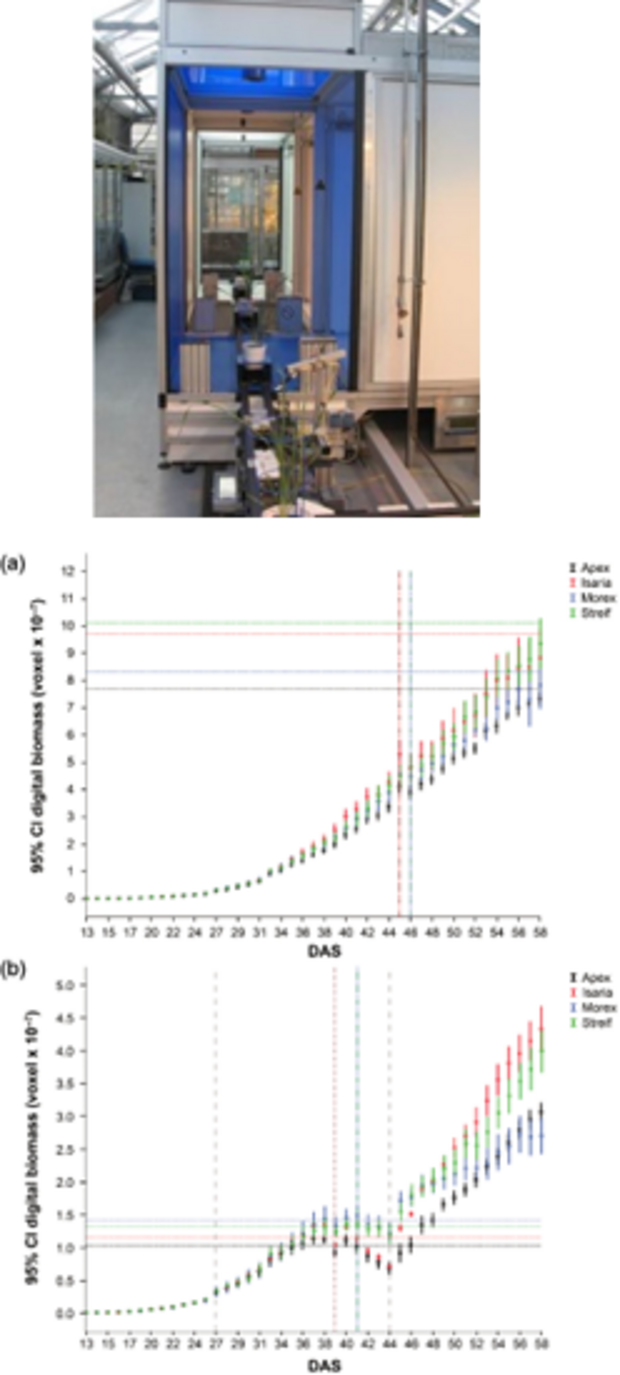
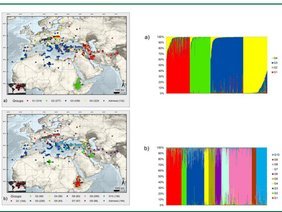
Our ability to leverage the genetic diversity maintained in ex situ collections is strongly dependent on the knowledge about genes that underlie the expression of phenotypic traits. Therefore, the overriding goal of the research group is the development of biological resources, molecular tools and strategies facilitating an improved conservation utilization of plant genetic resources. Research activities focus mainly on barley (Hordeum vulgare), wheat (Triticum aestivum) and Bean (Phaseolus vulgaris). DNA marker analysis and sequencing are being employed for the quantification of genetic variability and its relation to phenotypic variability. The corresponding data are being used for the development of core collections and populations to be used for association mapping (Fig. 1, 2). Moreover, fingerprinting data are evaluated to assess the genetic integrity of genebank accessions to develop a DNA marker based quality management of selected collections. Our research follows two main threads, improvement of the collection management and genetics of biomass accumulation, both of which contribute to IPK research theme Strategies for the Valorization of Genetic Resources.
Scientists of the research group are teaching at the Institute of Agricultural Sciences at Martin Luther University Halle-Wittenberg.