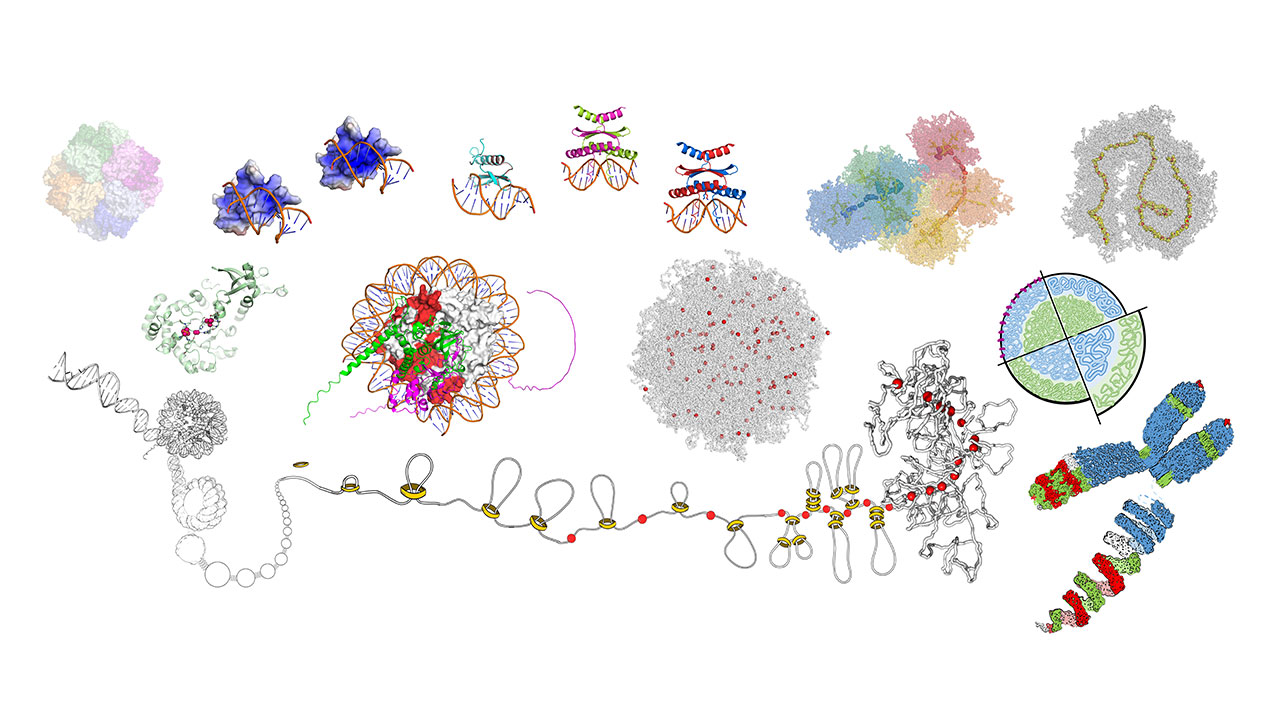
In silico Genebank Proteomics
“Everything that living things do can be explained by the jiggling and wiggling of atoms.” (Richard Feynman)
Proteins and nucleic acids are essential to molecular processes of life. They are both polymer biomolecules whose macro arrangements confer functions beyond of that of their smallest parts, amino acids and nucleotides, respectively.
With the current developments on genomics, much information is being generated on the linear sequences of amino acids and nucleotides. The task of our group is to translate this information into structural models of proteins and chromosomes. Such models are essential to understand how these macro molecules work and interact. Our goal is to find common and variable molecular features and mechanisms in different plant species, helping to elucidate how proteins and chromosomes structurally evolve and adapt to their changing environments.
The group contributes to IPK Research Theme “Genome Diversity and Evolution”.
scroll top
Projects
In Silico Genebank Proteomics
Proteins are the workforce of the cells. They are the link between the information guarded in the genome and the diverse phenotypes we observe. With the recent advances of artificial intelligence, we are now able to build structural models for all proteins of a species: its proteome. We leverage from AlphaFold’s recent development and the large number of accessions of crop plants in the Federal Ex situ Gene Bank. Our goal is to characterize the structural variability in variant proteomes, liking the sequenced genomes to the annotated phenotypes. Focusing first on barley, we are working to identify protein structural features that distinguish it from other species, and how much they change among barley variants. A consequence of such exploratory analyses is the construction of a database of structural annotations and predictions to each protein. Its main purpose is to allow comparative analyses between different proteins and to help predicting the effects of mutations. In collaboration with other groups of IPK, our models and inferences drawn from them can be exposed to experimental proof. Examples are transcriptional factors and proteins of the kinetochore complex.
Literature:
journals.plos.org/plosone/article?id=10.1371/journal.pone.0192826
Polymer models of chromosome dynamics
In the three-dimensional space, long DNA molecules look like yarn bundles. The way they pack changes during the cell cycle and varies between cell types and species. This variable organization facilitates cell division and gene regulation. In collaboration with experts in chromosome conformation capture sequencing, and in microscopy techniques that reach super resolution, we evince the spatial organization of chromosomes with unforeseen details.
The goal of this project is to provide polymer models of chromosomes that unravel the mechanisms driving conformational changes. With a large range of genome sizes, chromosome numbers and DNA repeats distribution, plants are the perfect showcase on how to modulate chromosome conformation.
Literature:
scroll top
Staff
scroll top
Publications
Šimková H, Câmara A S, Mascher M:
Hi-C techniques: from genome assemblies to transcription regulation. J. Exp. Bot. (2024) Epub ahead of print. https://dx.doi.org/10.1093/jxb/erae085
scroll top